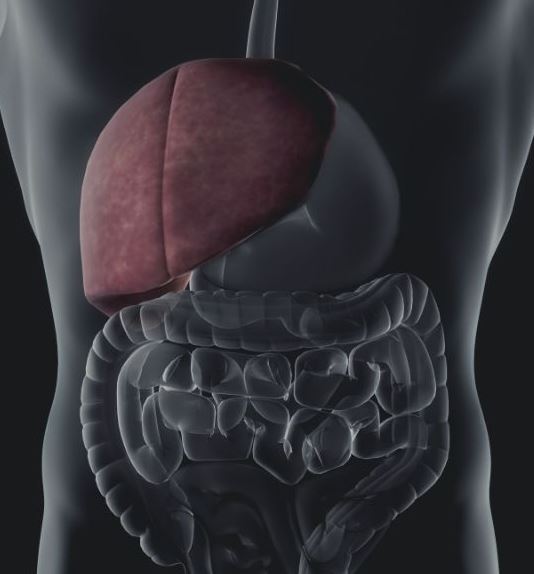
Hepatic stellate cells have recently gained a great deal of attention regarding their contribution to the progression of diseases such as liver fibrosis, non-alcoholic steatohepatitis (NASH), and hepatocellular carcinoma. They are mesenchymal cells that are located between sinusoidal endothelial cells
Gene Expression Profiling
-
发布: 六月 09, 2019完整阅读 »
-
发布: 八月 28, 2018完整阅读 »
qPCR is a powerful tool for quantification of gene expression levels and copy number variation. Despite the advances in next-generation sequencing (NGS), qPCR still serves as the "gold standard" for gene expression analysis. Due to poor reproducibility and vast lab-to-lab variation, all NGS data requires
-
完整阅读 »
Cancer immunotherapy is one promising cancer treatment option whereby the host's own immune system is used to treat cancer. The therapy works by either stimulating certain immune activities, or counteracting cancer cell signals that suppress immune responses. Cancer immunotherapy has progressed significantly
-
完整阅读 »
Aging and Telomere Length Quantification by qPCR
Aging is a time-dependent decline of the body’s functional capabilities and is an inevitable course of life (as shown in the image below, extracted from the Wall Street Journal). The rate of aging though is highly variable among individuals. When evaluating
-
完整阅读 »
The 2017 Nobel Prize in Physiology or Medicine was awarded to Jeffrey C Hall, Michael Rosbash, and Michael W Young for research that established key mechanistic principles on how circadian rhythms are regulated. Circadian rhythms are endogenous oscillations adjusted to changing external cues and driven
-
发布: 九月 20, 2017完整阅读 »
Next generation sequencing (NGS), such as Whole Exome Sequencing (WES) and RNA-Sequencing (RNA-Seq), provides a high throughput approach for DNA sequencing and gene expression analysis. It is rapidly advancing our knowledge on almost all aspects of genetic research and shedding light on how individual
-
发布: 可能 03, 2017完整阅读 »
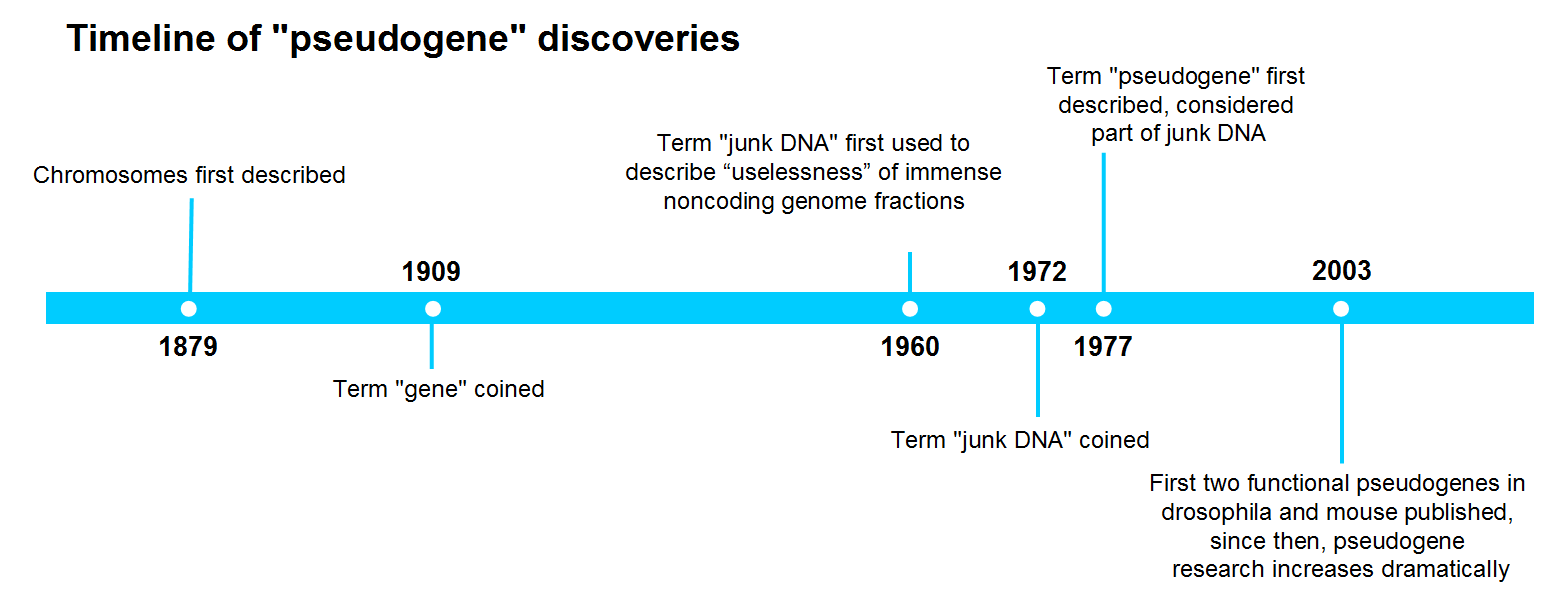
3 Reasons Why You Should Take "Pseudogenes" Seriously in Your Research
Protein-coding DNA, or “functional” genes, only accounts for up to 2% in the human genome. Until recently, the remaining 98% have been referred to as “junk DNA” for their putative noncoding feature. The name itself implies that junk
-
发布: 游行 26, 2017
-
发布: 二月 14, 2017完整阅读 »
SAN DIEGO, Calif. -- ScienCell Research Laboratories, Inc. recently announced it would be expanding its cell culture products to the gene expression profiling market. ScienCell, who prides itself on supplying unique primary cell types for nearly 20 years, will now supply qPCR array kits and individual
-
发布: 二月 14, 2017完整阅读 »
Online webinar hosted by sciencell to learn tips for successful qPCR, a technique for gene expression profiling
CARLSBAD, CALIFORNIA, USA, -- Sciencell Research Laboratories, Inc. , a global provider of primary cells, cell culture media and reagents for life science industry, announced today that it
-
发布: 十二月 06, 2016完整阅读 »
Cell-based assays are widely used in basic and translational research as cost-effective and accessible models to mimic in vivo responses. To obtain reliable data, assessing the health of cultured cells prior to any assays is essential. Furthermore, many cell-based assays require quantification of cell
-
发布: 八月 23, 2016完整阅读 »
In this final installment on how to improve your qPCR gene expression profiling (see 6 Tips to Improve qPCR: part 1 and part 2 for previous discussion), we will discuss pipetting tips and the benefits of a separate reverse transcription step in qPCR template preparation.
Tip #5. Good Laboratory Practice
-
发布: 八月 17, 2016完整阅读 »
After watching displays of astounding athletic prowess in the 2016 Olympics, I was inspired to take a closer look at the science behind exercise training, recovery, and injury with a focus on the importance of blood vessels during exercise.
Let’s start with some basic training: Why are blood vessels
-
完整阅读 »
Despite being essential for any experiment involving DNA replication, people rarely give primers a second thought. To coincide with the launch of our new qPCR gene reference tool, we’ll be giving them a little more of the praise they deserve.
Dr Kary Mullis received the 1993 Nobel Prize in Chemistry -
发布: 四月 26, 2016完整阅读 »
In this next installment on how to improve your qPCR gene profiling (see 6 Tips to Improve qPCR for previous discussion), we would like to discuss the limitations of melting curve analysis and PCR inhibitors.
Tip #3. Melting curve analysis is NOT sufficient for assessing qPCR product specificity.
In
-
发布: 二月 03, 2016完整阅读 »
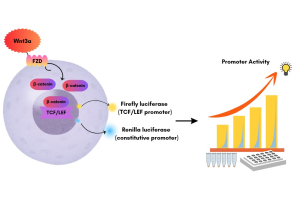
Gene expression analysis is critical for understanding the transcriptome profiles of primary cells and how they directly influence the cells’ functionality. In addition, the transcriptome profile can affect cell proliferation, differentiation, senescence, or apoptosis. Traditional reporter-gene assays
-
发布: 二月 03, 2016完整阅读 »
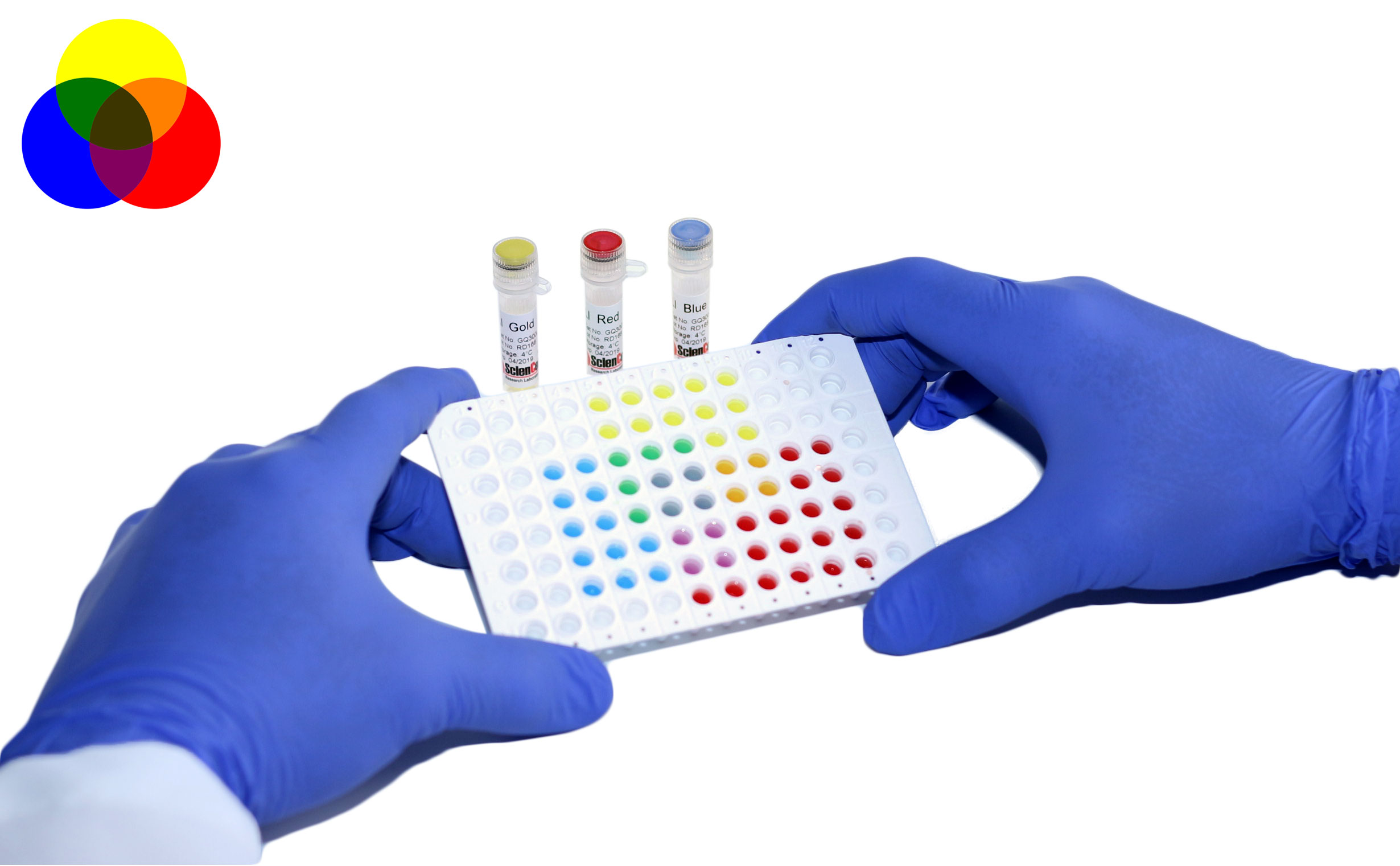
qPCR array provides a quick, powerful and sensitive approach for gene expression profiling. To help our customers acquire accurate and consistent results, we are happy to share some tips for better qPCR.
Tip #1. Good primer design is the key to a successful qPCR array.
The success of a qPCR array depends
-
发布: 十月 07, 2015完整阅读 »
Dye-based qPCR assays, such as SYBR® Green and EvaGreen®, have many advantages over probe-based qPCR assays. Dye-based systems are desirable because they are highly adaptable, cost effective, and time efficient. qPCR specificity, however, is a vital factor to consider when using dye-based qPCR assays.















.png)