Gene expression analysis is critical for understanding the transcriptome profiles of primary cells and how they directly influence the cells’ functionality. In addition, the transcriptome profile can affect cell proliferation, differentiation, senescence, or apoptosis. Traditional reporter-gene assays and cDNA microarrays often require either transfection of exogenous material or large quantities of high quality RNA. Primary cells, however, are notoriously difficult to transfect and the efficiency varies greatly between different cell types. In addition, primary cells have a finite lifespan and limited expansion capacity, making it difficult to obtain a high yield of RNA. To help address these challenges, ScienCell Research Laboratories has developed GeneQuery™ qPCR Array kits for gene expression profiling which we validated and optimized using primary cell cDNA. GeneQuery™ will enable researchers to directly measure protein expression patterns of primary cells, stem cells, tissues, and cell lines.
To quantify PCR progress in real time, we use a special fluorescent dye that only fluoresces when bound to double-stranded DNA. When it is free or when it is bound to single-stranded DNA, it is non-fluorescent. A good example of this type of dye is SYBR Green or EvaGreen. The fluorescent dye-based qPCR array provides a quicker and easier approach, with high specificity and reproducibility. Due to the high sensitivity and specificity of the assay, even genes with very low abundance can be analyzed.
GeneQuery™ is a highly adaptable research tool for studying the gene expression of cells and has many applications. First, GeneQuery™ can be used for basic research to study cell signaling, biological pathways and diseases. For example, our Human Cell Cycle kit (Cat# GK003) can be used to evaluate how genes expression changes will affect cell cycle progression. In addition, GeneQuery™ can be used to assist in drug screening to identify novel target genes, drug action mechanisms, and maybe even develop new drugs. Our GeneQuery™ Amyotrophic Lateral Sclerosis qPCR Array Kit (Cat# GK004), for example, can examine genes expression variations before and after drug treatment. Lastly, GeneQuery™ could be used to test drug efficacy and in the future may be applicable for personalized medicine because it could be used to predict if a drug will be effective for a particular patient. Studies for instance have shown that 5-FU, a common anticancer chemotherapy agent, shows better response if certain genes are differentially expressed upon treatment.
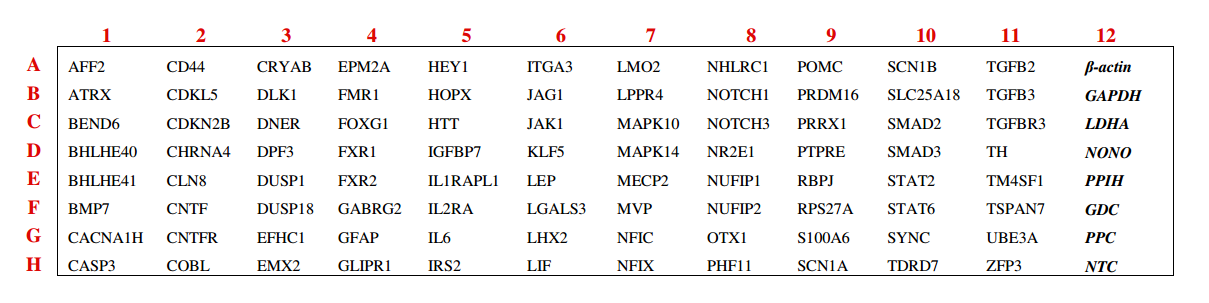
To demonstrate how powerful and convenient our GeneQuery™ arrays are, the GeneQuery™ Astrocyte Cell Biology kit (Cat# GK009) will be used as an example (figure 1). For this kit, 88 astrocyte cell biology related genes were selected to include well-characterized astrocyte markers, transcription factors related to regulating astrocyte differentiation and maintenance, various transcription factors regulating signaling pathways (including Notch, TGF-beta, JAK/STAT, and MAPK pathways), and genes relevant to astrocyte-associated diseases such as epilepsy, fragile X syndrome, Rett syndrome, and Alexander’s disease. Every GeneQuery™ kits includes 8 controls for quantification and quality control purposes. Five well-accepted target housekeeping genes, β-actin, GAPDH, LDHA, NONO, and PPIH are used for normalization of data. The additional 3 controls shown are internal controls. The Genomic DNA (gDNA) Control (GDC) detects possible gDNA contamination in the cDNA samples. The positive PCR Control (PPC) tests whether samples contain inhibitors or other factors that may negatively affect gene expression results. Finally, the No Template Control (NTC) contains only the β-actin primer set with no template, and it can be used to monitor the DNA contamination introduced during the workflow.
Using our own cDNA from normal primary human brain astrocytes compared to human pluripotent stems cells, we were able to acquire a large amount of information about these two cell types in only 2 hours. First, as expected we found that GFAP and CD44 (both astrocyte markers) were highly expressed in the human astrocytes but not detected in the pluripotent stem cells. Additionally, 4 other genes (HOPX, NR2E, PRRX1, and SYNC) were highly expressed in the astrocytes but not detected in the pluripotent stem cells. In brain astrocytes compared to HPSC, we also found 3 transcription factors that were upregulated more than 1,000 fold, 4 transcription factors upregulated more than 250 fold and 5 transcription factors that were downregulated. We also identified expression changes in transcription factors from the various signaling pathways such as MAPK, Notch, JAK/STAT and TGF-beta.
Additionally, using the GeneQuery™ human astrocyte cell biology kit, we compared human brain primary astrocytes (Cat# 1800) to human primary retinal astrocytes (Cat# 1870) (figure 2). We identified 5 region specific brain astrocyte genes expressed in brain astrocytes but not detected in retinal astrocytes. We also found several genes that are up-regulated specifically either in retinal astrocytes or in brain astrocytes. These region-specific genes may have implications in cell differentiation, biology functions, maintenance, and related signaling pathways. These are just two examples of how GeneQuery™ can be used to compare one cell type to another: one comparing mature cells to stem cells and one comparing the same cell type but from different locations. These kits could also be used to compare non-treated and treated primary cells to see how gene expression changes. Even though the results shown here are preliminary, you can see how quickly you can obtain relevant information from your cells and many new research avenues may be identified.
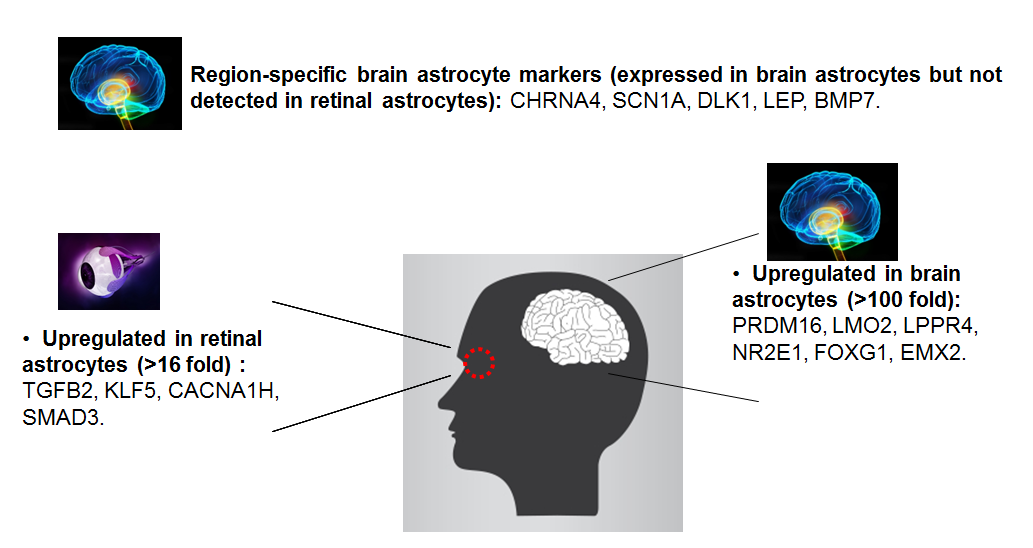
Figure 2. Preliminary results acquired by comparison of human brain astrocytes and human retinal astrocytes using GeneQuery™ Astrocyte Cell Biology Kit (Cat# GK009).
In addition to the pre-designed GeneQuery™ qPCR array kits, ScienCell Research Laboratories also provides individual primers sets for more than 1,000 genes, and custom primer design service. Please visit http://sciencellonline.com/genequery, or email info@sciencellonline.com for more information.
Images by Dream Designs from freedigitalphotos.net